In the field of genomic research, two powerful techniques—RNA sequencing (RNA-Seq) and CRISPR screening—are commonly used to study gene function, regulatory mechanisms, and disease processes. While both approaches provide valuable insights into the genome and its functionality, they differ significantly in their methodologies, applications, and the type of data they generate. Understanding the distinctions between RNA sequencing and CRISPR screening is crucial for researchers choosing the right approach for their experiments. This article will explore these two techniques in detail, highlighting their differences, advantages, and the roles they play in advancing genetic research.
What is RNA Sequencing (RNA-Seq)?
RNA sequencing, or RNA-Seq, is a high-throughput sequencing technique used to analyze the transcriptome—the complete set of RNA molecules transcribed from the DNA in a given cell or tissue type. RNA-Seq allows for the quantification of gene expression, detection of alternative splicing events, and identification of non-coding RNA molecules. By sequencing RNA, researchers can gain insights into how genes are regulated, how gene expression varies across different conditions, and how different RNA species contribute to cellular processes.
How RNA-Seq Works
- RNA Extraction: Total RNA is extracted from the sample of interest (e.g., a cell line, tissue sample, or organism).
- Library Preparation: RNA is reverse-transcribed into complementary DNA (cDNA) and then fragmented into smaller pieces for sequencing.
- Sequencing: The cDNA fragments are sequenced using next-generation sequencing (NGS) technologies.
- Data Analysis: The resulting sequences are mapped back to the reference genome or transcriptome to analyze gene expression levels, splicing patterns, and other features.
What Does RNA-Seq Provide?
- Gene Expression Profiles: RNA-Seq enables researchers to measure the abundance of transcripts from each gene, providing a snapshot of gene activity.
- Alternative Splicing Events: RNA-Seq can identify isoforms of a gene that arise due to alternative splicing, a key aspect of gene regulation.
- Non-Coding RNA: RNA-Seq also identifies and quantifies non-coding RNAs, such as microRNAs, long non-coding RNAs, and other regulatory RNA species.
What is CRISPR Screening?
CRISPR screening, on the other hand, is a functional genomics tool that leverages the CRISPR-Cas9 system to alter genes in a targeted manner. This technology allows for genome-wide or targeted perturbations of genes to study their function in various biological contexts. Unlike RNA-Seq, which focuses on measuring gene expression levels, CRISPR screening involves creating genetic modifications (such as gene knockouts, activations, or inhibitions) and analyzing how these modifications affect cellular behavior, disease processes, or responses to drugs.
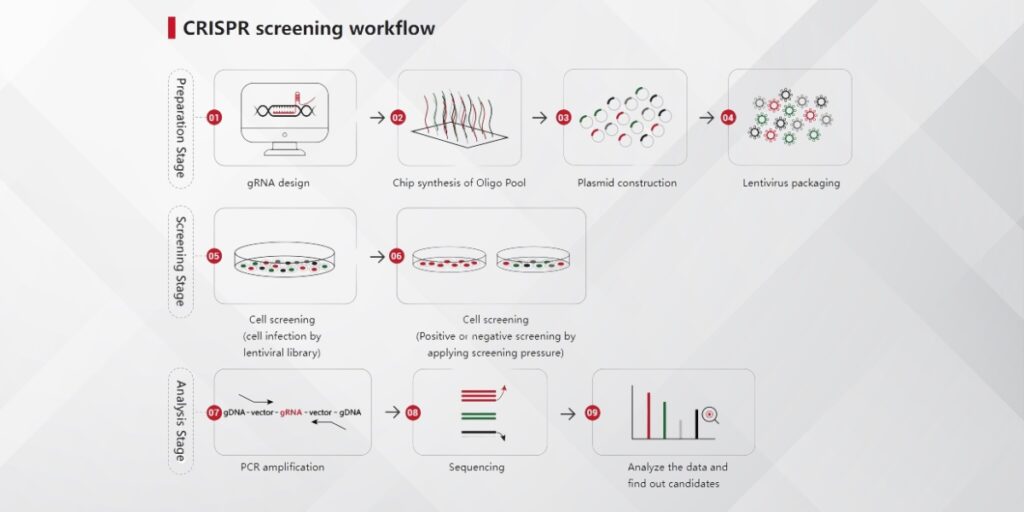
How CRISPR Screening Works
- Library Preparation: A CRISPR library is prepared, which contains a collection of single-guide RNAs (sgRNAs) designed to target different genes or specific gene families.
- Delivery: The CRISPR library is introduced into cells (via viral vectors or other delivery systems).
- Gene Editing: The CRISPR-Cas9 system edits genes by inducing double-strand breaks, leading to gene knockout, activation, or repression.
- Screening and Data Collection: Researchers screen for phenotypic changes (such as cell survival, proliferation, drug resistance, etc.) resulting from the gene modifications.
- Data Analysis: The results are analyzed to identify genes involved in specific processes, providing functional insights into their roles in biology.
What Does CRISPR Screening Provide?
- Gene Function Insights: By systematically knocking out or activating genes, CRISPR screening allows researchers to determine the functional roles of genes in specific cellular processes.
- Target Identification for Diseases: CRISPR screens are commonly used to identify genes that influence disease progression, drug resistance, and therapeutic responses.
- High-Throughput Functional Genomics: CRISPR screening can perturb thousands of genes in parallel, making it a powerful tool for large-scale genetic studies.
Key Differences Between RNA-Seq and CRISPR Screening
While both RNA-Seq and CRISPR screening provide valuable genomic insights, they serve different purposes and are applied in different contexts. Below are the main differences:
Purpose and Focus
- RNA-Seq: Focuses on measuring gene expression and understanding the transcriptome. It provides a snapshot of which genes are being transcribed and how gene expression levels vary under different conditions.
- CRISPR Screening: Focuses on gene function by perturbing genes and studying the phenotypic consequences. It is used to identify which genes are important for specific biological processes or disease mechanisms.
Methodology
- RNA-Seq: Involves sequencing RNA to analyze gene expression levels, splice variants, and non-coding RNA. It does not involve genetic modification of the genome.
- CRISPR Screening: Involves creating targeted genetic modifications in the genome, such as knockouts or activations, to study the functional consequences of gene perturbations.
Data Output
- RNA-Seq: Produces gene expression data, including transcript abundances, alternative splicing events, and regulatory RNA species.
- CRISPR Screening: Produces phenotypic data, including information about how gene modifications affect cellular behavior, disease phenotypes, or drug responses.
Applications
- RNA-Seq: Primarily used for studying gene expression patterns, alternative splicing, and regulatory RNA. It helps researchers understand the molecular changes in response to different conditions, including disease states.
- CRISPR Screening: Used to identify genes involved in disease mechanisms, drug resistance, and cellular processes. It helps researchers determine the functional role of genes by observing the effects of genetic modifications.
When to Use RNA-Seq vs. CRISPR Screening
- RNA-Seq is ideal for exploring the global expression of genes, understanding how genes are regulated, and identifying changes in gene expression due to disease, treatment, or environmental factors. If you are interested in measuring how genes are expressed in different conditions or identifying new transcript isoforms, RNA-Seq is the tool of choice.
- CRISPR Screening is suited for functional genomics studies where you want to understand the causal relationships between genes and phenotypes. If you’re looking to identify novel drug targets, study genetic vulnerabilities or map gene networks, CRISPR screening is the preferred approach.
Summary
RNA-Seq and CRISPR screening are complementary techniques that provide different but valuable insights into genomics. While RNA-Seq helps us understand the transcriptional landscape and gene regulation, CRISPR screening allows us to investigate gene function and discover causal links between genetic changes and phenotypic outcomes. Researchers often use these two techniques together to gain a deeper understanding of gene expression and function, which can drive advancements in drug discovery, disease modeling, and personalized medicine.
Both techniques play pivotal roles in modern genomics, and their complementary nature allows for a more comprehensive approach to understanding genetic networks and disease mechanisms. Depending on the research question, choosing between RNA-Seq and CRISPR screening—or using both—can provide powerful insights into the genetic basis of health and disease.
Published by Stephanie M.